
In January 2020, Caltech biochemist Pamela Bjorkman asked for volunteers to help work out the structures of immune proteins that attack a newly discovered coronavirus. The pathogen had emerged in China and was causing severe pneumonia-like symptoms in the people it infected. Knowing the molecular arrangements of these antibodies would be an important step toward developing drugs to fight the virus.
Christopher Barnes, a postdoc working in Bjorkman’s lab on the structure of HIV and the antibodies that target it, jumped at the chance to solve a new puzzle. “I was like, ‘Oh, I’ll do it!’” Barnes says. At the time he wasn’t aware how urgent the research would become.
Now, we are all too familiar with SARS-CoV-2, which causes COVID-19 and has killed more than 6 million people globally. Studies of the structure of the virus and the antibodies that target it have helped scientists quickly develop vaccines and treatments that have saved tens of millions of lives. But the virus continues to adapt, making changes to the spike protein that it uses to break into cells. That has left researchers scrambling for new drugs and updated vaccines.
Using high-resolution imaging techniques, Barnes is probing coronavirus spike proteins and the antibodies that attack them. His goal: Find a persistent weak spot and exploit it to create a vaccine that works against all coronaviruses.
Standout research
Barnes’ team used cryo-electron microscopy to reveal the structures of eight antibodies that stop the original version of SARS-CoV-2. The technique catches cells, viruses and proteins going about their business by flash freezing them. In this case, the team isolated coronavirus particles entwined with immune system proteins from people with COVID-19.
The antibodies had attached to four spots on the spike protein’s receptor binding domain, or RBD, the team reported in Nature in 2020. This fingerlike region anchors the virus to the cell it will infect. When antibodies bind to the RBD, the virus can no longer connect to the cell.
Plan of attack
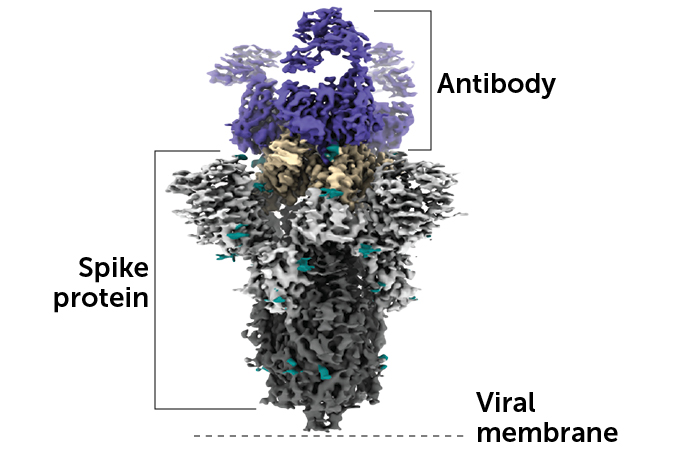
This image shows the 3-D molecular structure of an antibody latched onto a spike protein on the surface of the virus that causes COVID-19. The receptor binding domain is shown in tan.
 C.O. BarnesC.O. Barnes
C.O. BarnesC.O. Barnes
Barnes’ team also created an antibody classification system based on the RBD location where the immune system proteins tend to latch on. “That’s been really helpful for understanding the types of antibody responses that are elicited by natural infection,” says structural biologist Jason McLellan, who wasn’t involved in the work, and for identifying prime candidates for drug development.
“A major strength of Chris is that he does not limit himself or his research to one technique,” says McLellan, of the University of Texas at Austin. “He quickly adapts and incorporates new technologies to answer important questions in the field.”
Since launching his own lab at Stanford, Barnes and colleagues have determined the structures of six antibodies that attack the original SARS-CoV-2 virus and delta and omicron variants. Those variants are skilled at evading antibodies, including lab-made ones given to patients to treat COVID-19.
The newly identified antibodies, described in the June 14 Immunity, target the spike protein’s N-terminal domain. The structures of the sites where the proteins attach are the same in delta and omicron, hinting that the sites might remain unchanged even in future variants, the team says. Eventually, scientists may be able to mass-produce antibodies that target these sites for use in new therapies.
What’s next
Barnes has now turned his attention to antibodies that can fend off all coronaviruses — from ones that cause the common cold to ones found in livestock and other animals that have the potential to spill over into people.
Barnes and immunologist Davide Robbiani of the University of Lugano in Switzerland identified classes of antibodies that target variants from all four coronavirus families, blocking the viruses’ ability to fuse with cells.
What’s more, the structure of one of the binding sites on the spike protein is the same across the coronavirus family tree, Barnes says. “This is something you wouldn’t want to mutate as you diversify your viral family because this is a critical component of how you enter the cell.”
Two independent teams have identified similarly broad action in the same antibody classes. Taken together, the findings suggest that a universal coronavirus vaccine is possible, Barnes says.
“We’ve all kind of discovered this at the same time,” he says. The teams are now thinking, “Wow, this exists. So let’s try to make a real, true pan-coronavirus vaccine.”
Want to nominate someone for the next SN 10 list? Send their name, affiliation and a few sentences about them and their work to [email protected].

